Single-Point Wuestebach#
This test case at point scale covers the Wuestebach test site that is part of the TERENO network. Wuestebach is a forest site located in the Eifel region in Germany. To set up eCLM and run this test case, follow the instructions below.
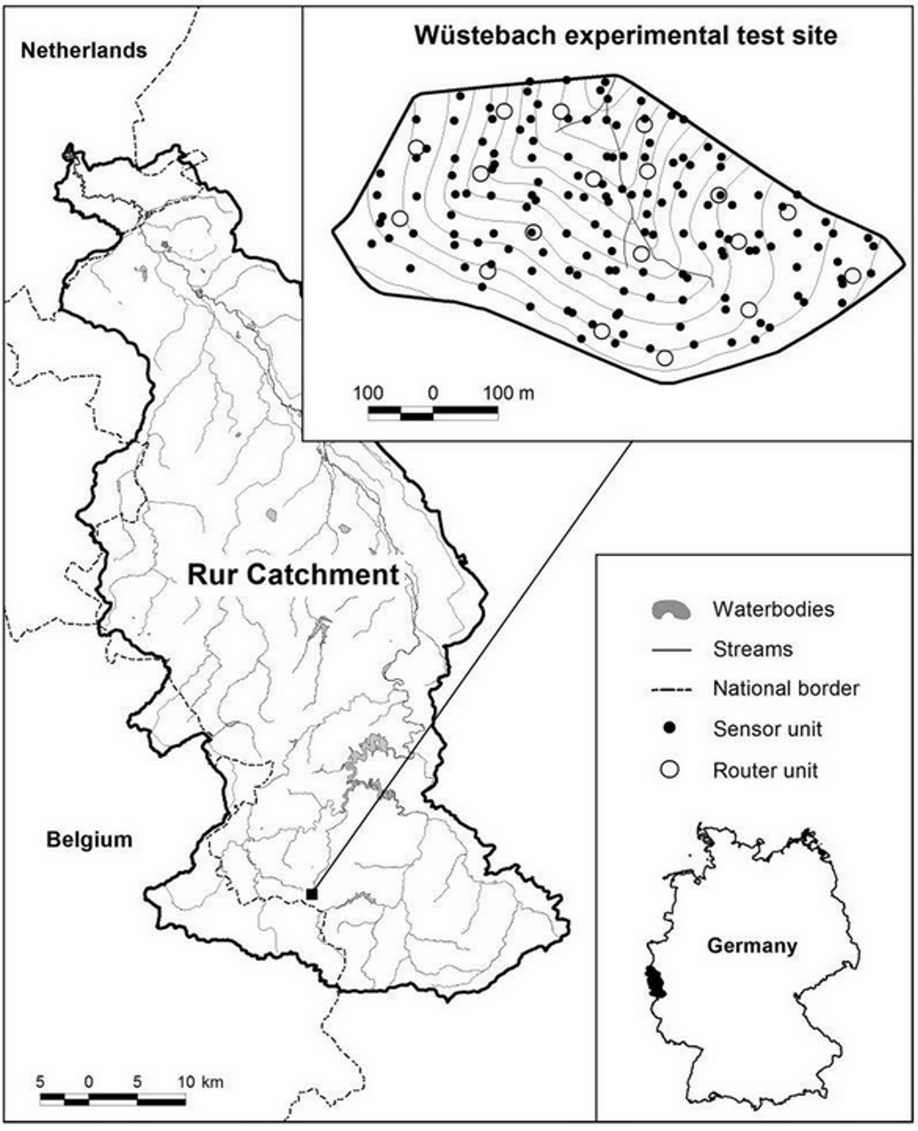
Fig. 3 Location of the Wüstebach test site within the TERENO Rur/Lower Rhine Valley observatory. Adapted from Bogena et al (2010).#
1. Copy the namelist files#
For JSC users, all required namelist and input files to run this case are in the shared directory /p/scratch/cslts/shared_data/rlmod_eCLM
mkdir test_cases
cp -r /p/scratch/cslts/shared_data/rlmod_eCLM/example_cases/wtb_1x1 test_cases/
cd test_cases/wtb_1x1
1. Download and extract data files (For non JSC users)#
You can download all required files through the JSC datahub.
mkdir -p test_cases/1x1_wuestebach
wget https://datapub.fz-juelich.de/slts/eclm/1x1_wuestebach.tar.gz
tar xf 1x1_wuestebach.tar.gz -C test_cases/1x1_wuestebach
The repository contains two directories. The common directory contains some general input files necessary to run eCLM cases. The wtb_1x1 directory contains the case specific domain and surface files as well as atmospheric forcing data and a script for namelist generation.
# Generate namelists
cd test_cases/1x1_wuestebach
export ECLM_SHARED_DATA=$(pwd)
cd wtb_1x1
clm5nl-gen wtb_1x1.toml
# Validate namelists
clm5nl-check .
2. Check the case setup#
You can check out the important namelist files.
The
lnd_infile is the primary configuration file for the land model component in CLM. It includes settings related to various processes and configurations specific to land surface modeling. For a complete list oflnd_in-namelist options, see https://docs.cesm.ucar.edu/models/cesm2/settings/current/clm5_0_nml.htmlThe
drv_infile configures the coupler and driver settings that control how different components of the Earth system model interact. It manages the synchronization and communication between components such as the atmosphere, land, ocean, and sea ice models. For a complete list ofdrv_in-namelist options, see https://docs.cesm.ucar.edu/models/cesm2/settings/current/drv_nml.html
Print out the namelist files and look at the customized configurations.
cat lnd_in
cat drv_in
3. Run the test case#
Customize the copied job script run-eclm-job.sh as desired. In this example, it is already customized to this test case, you should just adapt the SBATCH parameters --account to your compute project and --partition to your system. As Wuestebach is a single-column case, the number of processors should be set to 1 (SBATCH parameter --ntasks-per-node=1).
For non JSC users: Create a job script in your case directory with:
cat << EOF > run-eclm-job.sh
Adapt the SBATCH parameters and then copy the following in the shell file:
#!/usr/bin/env bash
#SBATCH --job-name=wtb_1x1
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --account=jibg36
#SBATCH --partition=batch
#SBATCH --time=1:00:00
#SBATCH --output=logs/%j.eclm.out
#SBATCH --error=logs/%j.eclm.err
ECLM_EXE=${eCLM_ROOT}/install/bin/eclm.exe
if [[ ! -f $ECLM_EXE || -z "$ECLM_EXE" ]]; then
echo "ERROR: eCLM executable '$ECLM_EXE' does not exist."
exit 1
fi
# Set PIO log files
if [[ -z $SLURM_JOB_ID || "$SLURM_JOB_ID" == " " ]]; then
LOGID=$(date +%Y-%m-%d_%H.%M.%S)
else
LOGID=$SLURM_JOB_ID
fi
mkdir -p logs timing/checkpoints
LOGDIR=$(realpath logs)
comps=(atm cpl esp glc ice lnd ocn rof wav)
for comp in ${comps[*]}; do
LOGFILE="$LOGID.comp_${comp}.log"
sed -i "s#diro.*#diro = \"$LOGDIR\"#" ${comp}_modelio.nml
sed -i "s#logfile.*#logfile = \"$LOGFILE\"#" ${comp}_modelio.nml
done
# Run model
srun $ECLM_EXE
EOF
Then you can submit your job:
sbatch run-eclm-job.sh
To check the job status, run sacct.
The model run is successful if the history files (wtb_1x1.clm2.h0.*.nc) have been generated.